Feature Story
Surer Signs of Life
If we ever think we’ve found signs of life elsewhere in the solar system, how would we convince ourselves that they’re real? A research team at APL is developing new palm-sized tools to help address that problem, targeting some of the strongest molecular signs of life, and even designing a way to sequence genetic material found on another world.
Jeremy Rehm
When they reached Mars’ surface in 1976, NASA’s two Viking landers touched down with a gentle thud. At 7 feet tall and 10 feet long and weighing around 1,300 pounds, these spacecraft — the first U.S. mission to successfully land on the Martian surface — looked like overgrown pill bugs.
What lay before them was a rusty, dusty wasteland littered with rocks under a tan-orange sky, far removed from the bustling alien metropolises science fiction writers and films had depicted. Scientists never expected alien cities, but they did suspect colonies of microbial aliens might be lurking in Martian soil. The landers were the first to search for extraterrestrial life.
Both landers were equipped with three automated life-detection instruments, each of which incubated a sample from the surface, studying the air above for molecules such as carbon dioxide, which could indicate photosynthesis, or methane, which microbes might produce as they metabolize nutrients the landers provided.
One of the instruments got a positive signal. The labeled release experiment, tracking radioactive carbon as it moved from digestible sugar to digested carbon dioxide, saw the telltale sign of living, metabolizing microbes.
The two other experiments, however, never did.
When NASA’s Viking landers imaged Mars’ surface, they showed a barren land of rocks and dust. Scientists suspected microbes might be living in that dust, however, and the landers were in part there to find them in the first ever life-detection mission on another planet.
Credit: NASA/Jet Propulsion Laboratory/Johns Hopkins APL
That maybe-discovery sparked a debate that persists even today, with proponents insisting (and new research suggesting) that only something alive could have made that positive signal.
But like many in the scientific community, Kate Craft, a planetary scientist at the Johns Hopkins Applied Physics Laboratory, remains skeptical. “It was a good experiment, but it was very limited in what it was able to detect,” she stated.
For one, the Viking experiments assumed microbes on Mars would eat the nutrients we provided them, which isn’t necessarily true. And even if they did, it’s still hard to believe just one line of evidence. “We always want to have positives on multiple signatures,” she said.
More problematic, however, is that scientists at the time didn’t know Mars’ surface is covered in perchlorate salts, minerals containing chlorine and oxygen that experiments show can destroy organic molecules and microbes when heated — producing chlorine gases, which the Viking landers in fact did detect. Nobody knew the salts were there until 2008 when NASA’s Phoenix lander discovered them.
For Craft and her colleague Chris Bradburne, a biologist and senior scientist at APL, the Viking missions underscored the monstrous challenge scientists face to definitively say we’ve found life on another world. The type, surety and repeatability of that evidence all matter. Numerous spacecraft since the Viking landers have returned to Mars, searching for organic molecules, which contain mostly carbon, hydrogen and oxygen. They’re commonly associated with life but not sure indicators of it.
But the revelation about salts on Mars highlighted a more salient, albeit somewhat uninspiring, point: The chances of detecting signs of life with even the best technology are likely slim if you don’t purify your samples first.
It’s much sexier to think about the detector. But if you can’t prep your samples and optimize them so your sensor can detect what you’re after, they don’t do you any good.
Researchers have fixated on the detection side of the equation, but the sample preparation — an earlier step in the workflow — has gone mostly ignored. Salts are particularly worrying because they can make analysis difficult, and the prime targets for future life-detection missions are places with salty, liquid water oceans beneath their surfaces — worlds like Jupiter’s moon Europa and Saturn’s moon Enceladus.
Since 2013, Bradburne, Craft and a team of researchers at APL have been developing new, palm-sized microfluidic systems for future spacecraft to address that challenge. They can purify and isolate molecules that could be strong indicators of life — amino acids, proteins, RNA, DNA.
“It’s much sexier to think about the detector,” Bradburne said. “But if you can’t prep your samples and optimize them so your sensor can detect what you’re after, they don’t do you any good.”
But the team is pushing one of their instruments even further: a sequencer for space. It would not only prep and concentrate long-chain molecules like DNA and RNA but also pump out their entire genetic code right at the destination. Additionally, it would detect these molecules whether they’re like terrestrial DNA and RNA or not, providing the ability to detect life with an entirely separate origin.
“It could give you a really conclusive signal,” Bradburne said. You just have to figure out how to build it.
The Cleaning Machines
Craft and Bradburne had considered creating a sample preparation chip for DNA and RNA back in 2014, building off work Bradburne started a few years earlier.
As far as life indicators go, DNA and RNA sit relatively high on the list because both form the backbone from which all Earth life has evolved. But it’s for that exact reason that many scientists were skeptical of searching for DNA and RNA elsewhere in the solar system.
For genetic material to pass down information between generations, they argued, organisms would already have had to evolve to some extent — a fairly unlikely possibility, Craft said. As such, many scientists considered DNA and RNA less important biosignatures and instead prioritized life’s other building blocks, such as amino acids — the constituents of all proteins and enzymes. “Life wouldn’t have to be ‘as evolved’ for those signatures,” Craft explained.
So, the team switched gears to make a miniature sample preparation system for amino acids. APL chemist Jen Skerritt, chemical engineer Tess Van Volkenburg and, later, Korine Ohiri, an expert in microfluidics, joined the team. Since 2018, they’ve been gradually perfecting the design.
At about 4 inches wide, 4 inches long and 2 inches tall, the system can easily fit in the palm of your hand. Yet it’s equipped with all the pumps and valves needed to push a sample through. The active region of the latest design is filled with tiny beads that attract amino acids in acidic solutions while salts and other gunk continue to flow out the other side into a waste deposit. After the sample passes through, the amino acids are stripped from the beads with a basic solution and shipped to whatever detector is attached to the chip.
Designing a prep system for space hasn’t been easy, Ohiri said. The amount of available power is fractions of what can be used in the lab, and the materials need to withstand potentially extreme temperatures and radiation. The team is currently making the amino acid purification system from common rapid prototyping materials, such as high-resolution resins used in 3D printing, but getting the material to be space-worthy while maintaining its performance, Ohiri said, remains challenging. “But that’s what’s so exciting about this project: There are so many aspects that are really at the leading edge.”
The trade-off with amino acids, however, is that they’re everywhere — from meteorites to comets to interstellar clouds. Certain clues can indicate whether they’re biological or not. Amino acids come in two forms that are mirror images of each other: one considered left-handed, the other right-handed. Through some fluke of evolution, all life on Earth uses just the left-handed amino acids. So by extension, if one type appears more than the other in a sample from another world, it could be a sign of life.
Bradburne, however, doesn’t completely buy it. “How do you know it’s not just contamination?” he asked, such as from a hitchhiking microbe that somehow escaped the deep cleaning process all spacecraft go through before launch. Detecting life in the universe, he says, comes down to not just detecting the molecules you’re looking for, but minimizing the chances of getting a false positive and making sure your experiments are repeatable.
DNA and RNA aren’t necessarily better for addressing those problems, unless you can sequence them. And that’s why, when nanopore sequencers were invented, the team saw a novel opportunity.
The Road to Sequencing
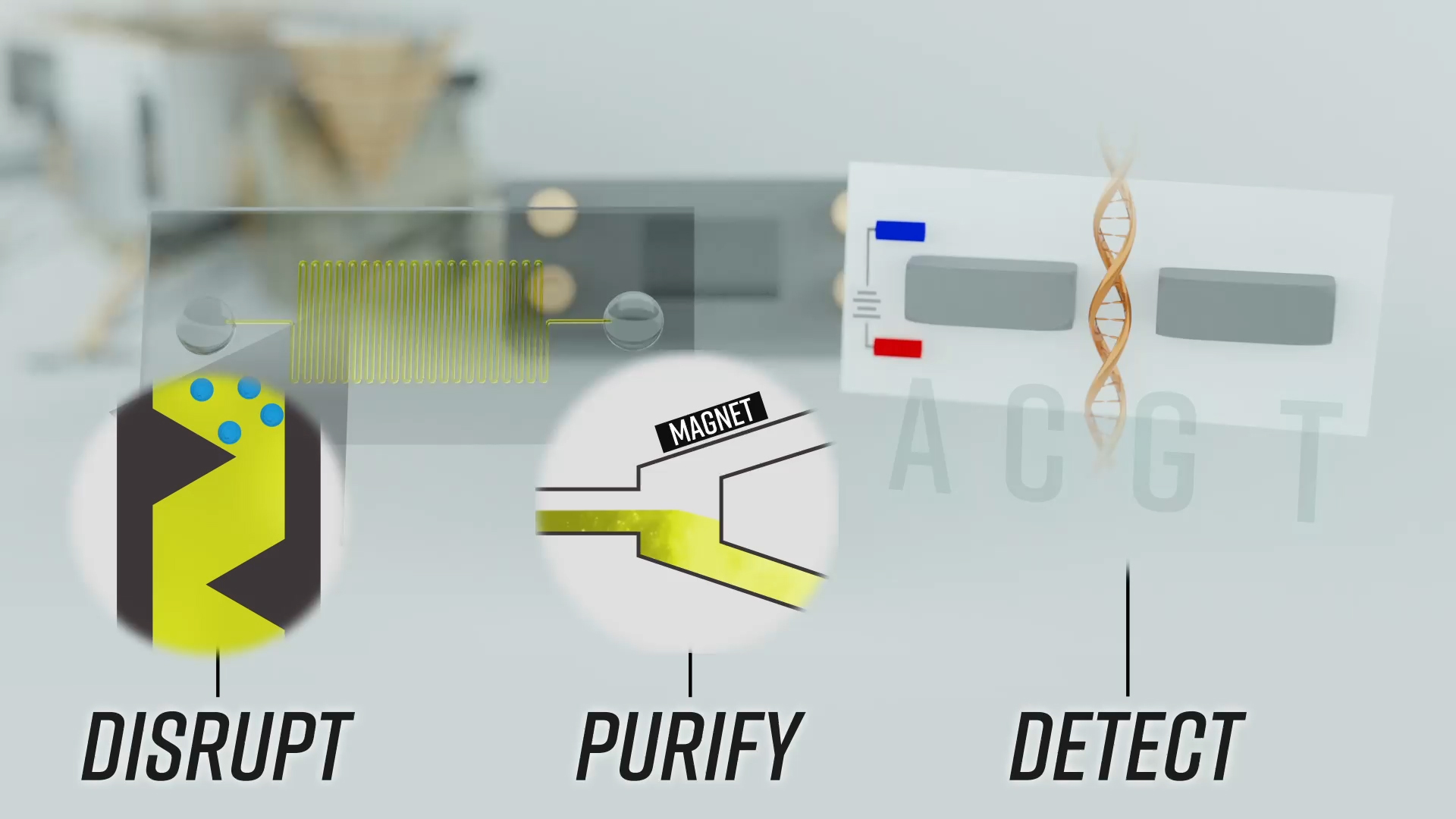
Nanopore sequencers are small, thumb-drive-size machines that can take a strand of DNA or RNA and read out the series of molecular building blocks that it’s made of. The strand moves through a pore that’s just billionths of an inch wide and that has an electric field passing through it. Each nucleotide uniquely disrupts that electric field as it moves through the pore. And a computer can interpret that disruption and say exactly which nucleotide just passed through.
How to isolate and sequence DNA in space: Start with a disruption phase, using sound or other waves to pulse magnetically attractive beads so they crack open spores or cells and let the DNA out. The DNA attaches to the beads, which are then pulled toward a magnet during the purification step. The beads are then washed to remove the DNA, which is then sent to a nanopore sequencer. The sequencer then reads out the chain of molecules that make up DNA — C, A, T and G. This setup should theoretically work for any long-chain molecule like DNA, including RNA, proteins or something entirely new.
Credit: Johns Hopkins APL
Besides being the ideal size for a spacecraft, Bradburne said, nanopore sequencers should, in theory, be able to interpret any type of long-chain molecule that comes through — DNA, RNA, proteins or some unknown XNA. But they also shrink the chances that a signal isn’t just a stowaway microbe. Earth-stemmed organisms have recognizable strands, such as those that code for specific enzymes and other proteins common to living things on Earth. So if sequences seem to match those frequently found here on Earth, they’re likely a false positive.
“The scientific returns would just be amazing,” Bradburne said.
There are a slew of reasons, however, why current nanopore sequencers aren’t ready for space. For one, they’re made of materials that can’t withstand years of subfreezing temperatures and radiation; even on Earth, they only last about six months. Even more problematic is that they use proteins from staph bacteria for the pore, raising concern about accidentally introducing biological products from Earth.
Those challenges have forced the team to instead start developing its own sequencer and accompanying sample preparation system.
“The idea is that, eventually, we’ll have a full instrument to prepare the sample the way we want it and then analyze it,” Craft said.
Enceladus is an ocean world where we have enough data to go beyond asking if it’s habitable. At Enceladus, we are ready to take the next step and search for signs of life.
The sample preparation component has made significant headway over the last year. The team is trying sound waves and other disruptive methods to break open cells and spores that may house the genetic material and magnetic beads to then hold onto the long-chain molecules.
But designing the nanopore sequencer has been more challenging. A synthetic platform with nanopores pressed into it is the most ideal, but how to control the pores’ size and make them so they slow the molecule so the computer can register each molecule in the chain as it passes through remains uncertain. A Canadian collaborator even suggested making the pores when they reach the destination to mitigate issues with shelf life. “I’m not sure how we’d do that, but nothing’s off the table right now,” Bradburne said.
Despite the obstacles, the team has wasted no time in talking about their tool with researchers developing concept missions. “We talk it up when we can,” Craft said, mostly to let people know that it’s an upcoming, viable instrument.
And one recent concept, a mission to Saturn’s moon Enceladus, includes something very similar to it.
Another Search for Life
At 314 miles wide — about the width of Pennsylvania — and on average nine times farther from the Sun than Earth, Enceladus should have been just a frozen ball of ice.
But in 2006, NASA’s Cassini mission revealed a tantalizing discovery: a plume of water vapor and ice spewing from four cavernous “tiger stripes” at Enceladus’ south pole. Various measurements indicate the faults link directly to a global liquid water ocean beneath the surface. The ocean may be interacting with the moon’s rocky core in a way similar to Earth’s deep-sea hydrothermal vents, where nearly 600 animal species live and thrive.
As Cassini passed through the plumes, it found molecules such as methane, carbon dioxide and ammonia — suspected chemical fragments of more complex molecules with four of the six elements key to life: carbon, hydrogen, nitrogen and oxygen.
“Enceladus is an ocean world where we have enough data to go beyond asking if it’s habitable,” said Shannon MacKenzie, a planetary scientist at APL. “At Enceladus, we are ready to take the next step and search for signs of life.”
MacKenzie recently led the development of a mission concept that would do just that. It’s called the Enceladus Orbilander, and it would operate just how it sounds: part-orbiter, part-lander. Six instruments would conduct measurements on material gathered from Enceladus’ plume to search for several potential biosignatures — left- and right-handed amino acids, fats and other long-chained hydrocarbons, molecules capable of storing genetic information, and even cell-like structures.
As a mission concept, the Orbilander study doesn’t identify specific instrument implementations like those that Craft and Bradburne’s team is producing, but it does include their conceptual ideas.
A plume of water vapor and ice particles spewing from the south pole of Saturn’s little moon Enceladus. When NASA’s Cassini spacecraft discovered the plume, it set off a series of subsequent finds that put Enceladus at the top of the list of places to search for life.
Credit: Johns Hopkins APL
“There’s always going to be some amount of uncertainty in search-for-life measurements,” MacKenzie said. “That’s why having a good sample prep step, which helps minimize the limit of detection, is so important, and why having instruments like the nanopore sequencer, which can offer both identification and characterization, is so critical.”
With the chance of sampling an ocean moon, Craft and Bradburne’s team is trying to determine how much water is needed to detect those biosignatures. And of course, it’s not easy. “I thought that we could go to these ocean worlds, dip our toes in and be able to see if life is there or not,” Craft said. But as she’s read research by oceanographers, she’s learned they have to filter liters of water to look for evidence of life — even here on Earth. “It’s just amazing. Because of all that water out there, it’s so dilute,” she said.
How do you collect such large volumes of water and concentrate them on another world? How do you process them in a microchip and see whether there are any important molecules there?
“There are just a bunch of challenges that haven’t been addressed yet,” Craft said. The team keeps plugging away though. Last month, they performed some experiments flushing various volumes of dilute amino acid samples spiked in ocean water through their sample chip. Initial results are promising, with the system capturing all amino acids at a range of efficiencies that will be reported in an upcoming science paper.
If ever moved from concept to launchpad, Enceladus Orbilander wouldn’t lift off until the mid-2030s, giving Craft and Bradburne’s team some time to further develop its tools. But even if the technology isn’t ready for that mission, Ohiri, like others on the team, remains optimistic that the technology will one day fly.
“My hope is that by the time the technology is mature enough, there will be a mission on the books, and we’ll be ready for it,” she said.
Areas of Impact
The Applied Physics Laboratory, a not-for-profit division of The Johns Hopkins University, meets critical national challenges through the innovative application of science and technology. For more information, visit www.jhuapl.edu.